2021. 2. 6. 02:09ㆍEnglish/Machine learning algorithm
Handle missing data
- Missing data can be addressed in two main ways:
1. Delete a sample (Row) or column (Feature) with missing data.
2. Use interpolation to predict missing data.
- Average, median, frequency, constant, etc. are used.
Let's use the code to check.
0. Create a data frame with missing data
Calling up required libraries
import pandas as pd # When it used to make DataFrame
import numpy as np # To calculate vector
from sklearn.impute import SimpleImputer # interpolation techniques
from io import StringIO # Leverage when creating data framesCreate a data frame with missing data
... """A,B,C,D
... 1.0,2.0,3.0,4.0
... 5.0,6.0,,8.0
... 10.0,11.0,12.0"""
df = pd.read_csv(StringIO(csv_data))
df
Data frames can be converted to a type of overflow array using 'dataframe.values'.
# We always get Numpy array from Pandas Dataframe
df.values
To determine which data is missing by column
df.isnull().sum()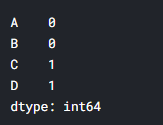
It can be seen that there is one missing data in columns C and D.
[1] Deleting a sample or column
# Delete Samples
df.dropna(axis = 0)
# Delete Columns
df.dropna(axis = 1)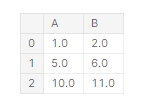
If you give Axis a value of 0, it will be deleted by record, and if Axis will be given a value of 1, it will be deleted by column.
This is also necessary knowledge for many functions using data frames, so it is better to remember.
# Parameters of 'dropna' method.
## When there are NaN values in all column, delete samples.
df.dropna(how='all')
The dropna function "how = 'all'" is a parameter that is deleted when all values are missing in a single sample.
Samples without all information are not needed, so it is better to delete them.
## When numbers of values is less than certain standard point, delete samples.
df.dropna(thresh=4)
thr is a parameter that sets the criteria for deletion.
The above expression gives a value of 4, which means that samples with less than 4 values should be deleted.
## When there are NaN values in certain columns, delete samples.
df.dropna(subset = ['C'])
A subset is a parameter that requires a sample to be deleted if a missing value exists in a particular column.
[2] Predicting missing data using interpolation
- Interpolation techniques are available in a variety of ways, but a separate library is provided for simplified use of interpolation techniques in a psychedelic run.
Mean
# We have to input data as numpy array, not pandas dataframe
## mean by columns
imr = SimpleImputer(missing_values = np.nan, strategy = 'mean')
imr = imr.fit(df.values)
imputed_data = imr.transform(df.values)
imputed_data
One of the main features of the cycle run is the use of estimator APIs. The main methods of this estimator are fit and transform. Fit is used to train the model and transform the data into parameters learned using transform.
It is important to note that the array of data to be transformed must equal the number of characteristics of the data used to learn the model. Some understanding of the estimator should be given, as it is not only used in interpolation, but also in the use of a psychedelic model.
* Additionally, it can be used at once through fit_transform.
When you enter missing_values and which interpolation techniques to use, it automatically adds missing data based on columns.
Median
## median by columns
imr = SimpleImputer(missing_values = np.nan, strategy = 'median')
imr = imr.fit(df.values)
imputed_data = imr.transform(df.values)
imputed_data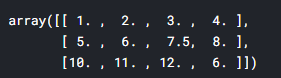
Frequency
## most frequent by columns
### It is imputated by first value in such columns when number of frequency of values is same.
imr = SimpleImputer(missing_values = np.nan, strategy = 'most_frequent')
imr = imr.fit(df.values)
imputed_data = imr.transform(df.values)
imputed_data
* If the frequency is the same, it is replaced based on the first value.
Even if a psychedelic estimator is not used, the following methods can be used to replace the missing data:
# It can be worked by this way
## mean by columns
imputed_data = df.copy()
imputed_data['C'] = df['C'].fillna(df['C'].mean())
imputed_data['D'] = df['D'].fillna(df['D'].mean())
imputed_data
In that case, the shape of the data frame remains the same.
* For your information, I use this method.
If you want to enter a value based on a row, you should use a somewhat complex method.
Frequency
# If you want to imputate by samples, you can do like this
from sklearn.preprocessing import FunctionTransformer
ftr_imr = FunctionTransformer(lambda x: imr.fit_transform(x.T).T, validate = False)
imputed_data = ftr_imr.fit_transform(df.values)
imputed_data
T is a transpose of a dimension from vector operations to a transpose that easily repositiones rows and columns.
It seems a bit complicated, so I wonder if it is necessary to do it in the following way. So I use the method below.
# You can also handle NaN data by samples like this
imputed_data = df.copy()
imputed_data.iloc[1] = imputed_data.iloc[1].fillna(imputed_data.iloc[1].mean())
imputed_data.iloc[2] = imputed_data.iloc[2].fillna(imputed_data.iloc[2].mean())
imputed_data

iloc is a method for finding values based on rows in a data frame.
In the example, the number of data is very small, so if there is a lot of data, you can use a statement or apply to handle the missing data.
For your information, if there is a lot of data, the for statement will take a very long time, so 'apply(lambda x:x.fillna(x.mean()))' can be solved quickly.
'English > Machine learning algorithm' 카테고리의 다른 글
| [8] Understanding K-Neighborhood (KNN) (0) | 2021.01.17 |
|---|---|
| [7] About RandomForest (0) | 2021.01.07 |
| [6] Information gain and impurity of decision tree (0) | 2021.01.06 |
| [5] Non-linear Troubleshooting with Kernel SVM (0) | 2021.01.04 |
| [4] Understand Support Vector Machine (SVM) Principles (linear) (0) | 2020.12.31 |